As computer vision and medical imaging is by its definition visual. I believe it’s really useful to make imaging research as interactive and accessible as possible.
This could be by providing additional code, data or some other form of interactivity that gives people a feel for the results.
Github is obviously great for code sharing, but what’s the easiest way of making interactive examples available on the web?
What works for me…
3D visualisation using x3dom with a static web page.
x3dom provides a nice markup language for including 3D rendering in html pages. This allows interactive rotation, zoom, scroll as well as setting up animations.
This demo here can be rotated and scrolled to view an animated mesh, which is produced from matlab using this script.
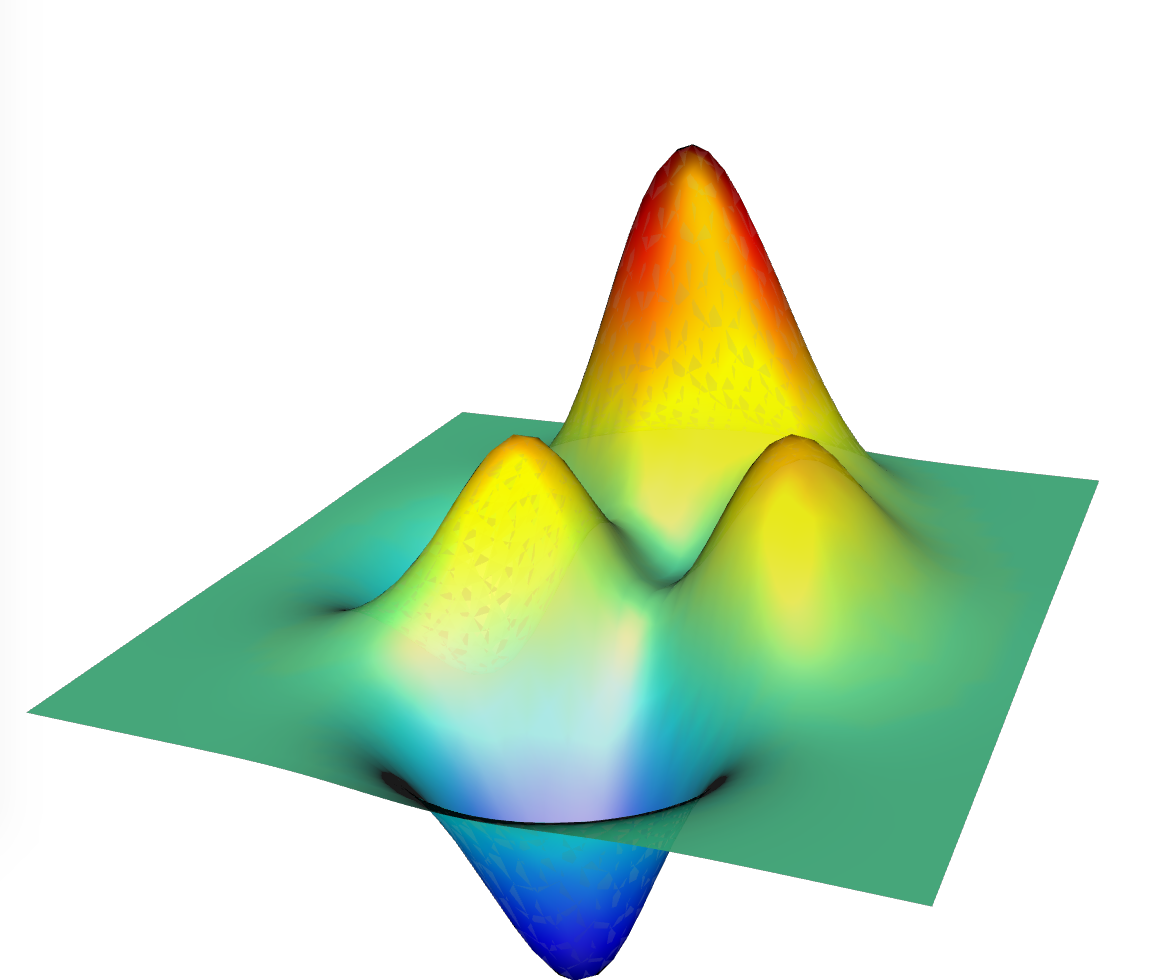
A number of 3D mesh libraries and software packages provide a way of converting a mesh or visualisation into x3d/x3dom. In python’s mayavi, figures can be saved as x3d and then put into html as x3dom.
I’ve written a few small Matlab plotting functions to plot normal and animated 3D meshes.
Throw in a bit of javascript/jquery and you can also manipulate the meshes in the browser.
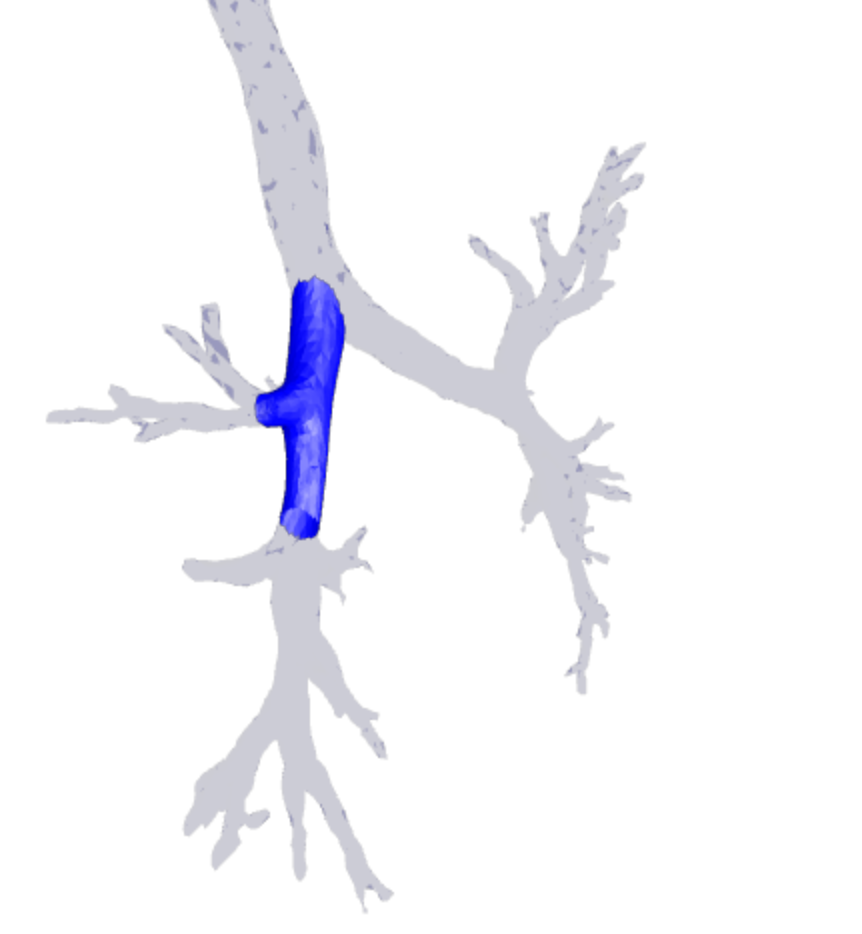
flask server for image processing
Sometimes a static page is not enough, and you want a server running that does some type of analysis/processing in the background.
I’m a big python user and I found flask to be a very simple way of creating a web server. This works well with standard visualisation libraries such as matplotlib or bokeh.

This example is hosted on heroku but there are many other PaaS that can be used.
Jupyter notebooks for interactive code
Jupyter notebooks are also great way of combining interactive code and plots but require the to run the notebooks locally to get all the benefits of the interactivity.